DECODE: A Deep-learning Framework for Condensing Enhancers and Refining Boundaries with Large-scale Functional Assays
by Zhanlin Chen, Jing Zhang, Jason Liu, Yi Dai, Donghoon Lee, Martin Renqiang Min, Min Xu, Mark Gerstein
Posted on January 27, 2021
Abstract
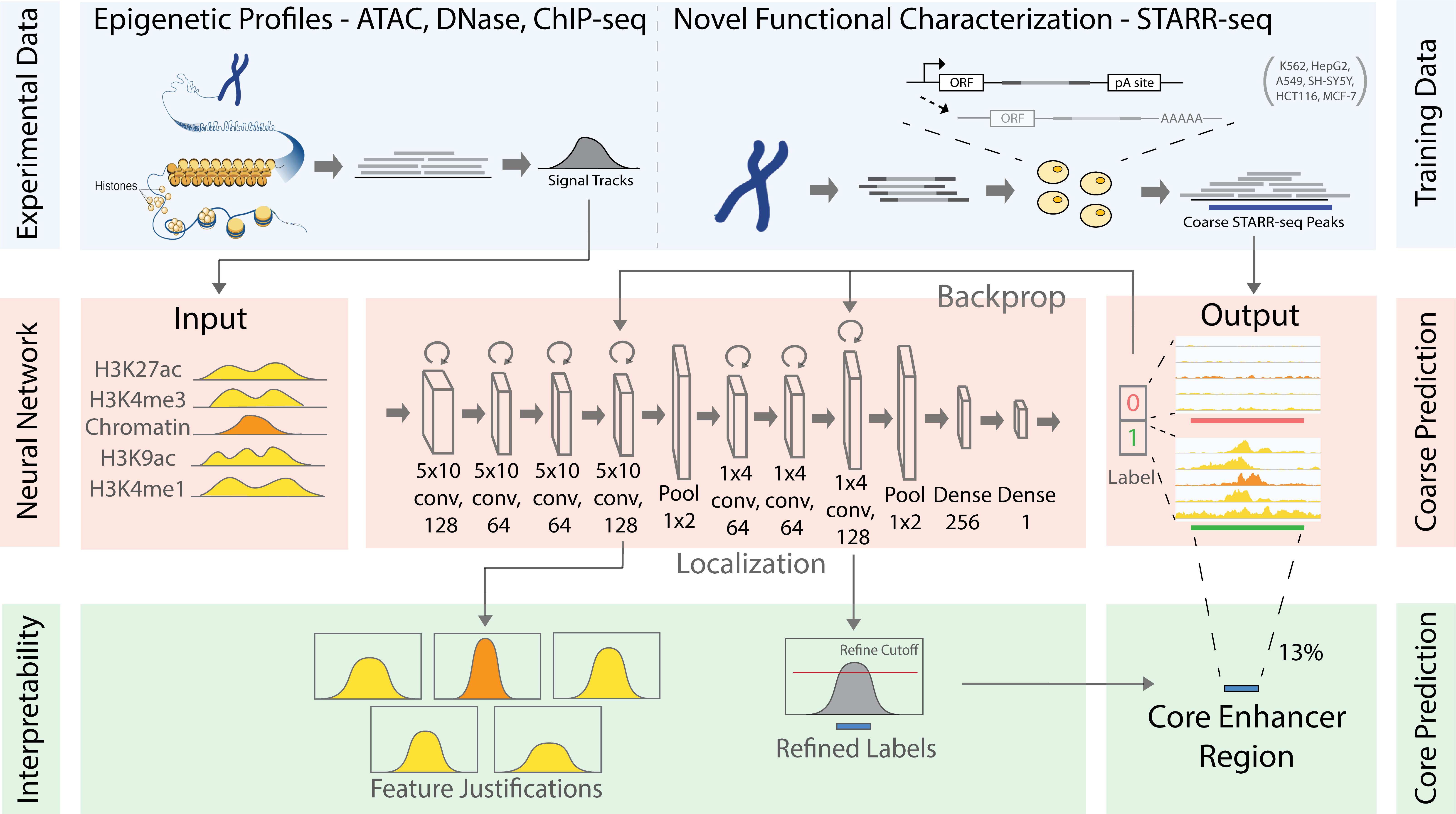
Summary: Mapping distal regulatory elements, such as enhancers, is the cornerstone for investigating genome evolution, understanding critical biological functions, and ultimately elucidating how genetic var-iations may influence diseases. Previous enhancer prediction methods have used either unsupervised approaches or supervised methods with limited training data. Moreover, past approaches have opera-tionalized enhancer discovery as a binary classification problem without accurate enhancer boundary detection, producing low-resolution annotations with redundant regions and reducing the statistical power for downstream analyses (e.g., causal variant mapping and functional validations). Here, we addressed these challenges via a two-step model called DECODE. First, we employed direct enhancer activity readouts from novel functional characterization assays, such as STARR-seq, to train a deep neural net-work classifier for accurate cell-type-specific enhancer prediction. Second, to improve the annotation resolution (~500 bp), we implemented a weakly-supervised object detection framework for enhancer local-ization with precise boundary detection (at 10 bp resolution) using gradient-weighted class activation mapping.
Results: Our DECODE binary classifier outperformed the state-of-the-art enhancer prediction methods by 24% in transgenic mouse validation. Further, DECODE object detection can condense enhancer an-notations to only 12.6% of the original size, while still reporting higher conservation scores and genome-wide association study variant enrichments. Overall, DECODE improves the efficiency of regulatory ele-ment mapping with graphic processing units for deep-learning applications and is a powerful tool for en-hancer prediction and boundary localization.
Contact:pi at gersteinlab.org